Short Notes on the State of Art and Challenges on Data Mining
Abstruse
3-dimensional (3D) structures of protein complexes provide fundamental data to decipher biological processes at the molecular scale. The vast corporeality of experimentally and computationally resolved protein-protein interfaces (PPIs) offers the possibility of grooming deep learning models to aid the predictions of their biological relevance. We present hither DeepRank, a full general, configurable deep learning framework for data mining PPIs using 3D convolutional neural networks (CNNs). DeepRank maps features of PPIs onto 3D grids and trains a user-specified CNN on these 3D grids. DeepRank allows for efficient training of 3D CNNs with data sets containing millions of PPIs and supports both classification and regression. We demonstrate the performance of DeepRank on two singled-out challenges: The classification of biological versus crystallographic PPIs, and the ranking of docking models. For both problems DeepRank is competitive with, or outperforms, state-of-the-art methods, demonstrating the versatility of the framework for research in structural biological science.
Introduction
Highly-regulated protein-poly peptide interaction networks orchestrate virtually cellular processes, ranging from DNA replications to viral invasion and immune defense. Proteins interact with each other and other biomolecules in specific ways. Gaining noesis on how those biomolecules interact in 3D infinite is key for understanding their functions and exploiting or engineering science these molecules for a wide variety of purposes such every bit drug design1, immunotherapyii, or designing novel proteinsiii.
In the past decades, a diversity of experimental methods (e.chiliad., X-ray crystallography, nuclear magnetic resonance, cryogenic electron microscopy) have determined and accumulated a large number of atomic-resolution 3D structures of protein-protein complexes (>7000 non-redundant structures in the PDBe databank (https://www.ebi.ac.uk/pdbe/) as of Sep. second 2020). Numerous automobile learning methods4, and recently several deep learning techniquesfive,6,7, take been developed to larn complicated interaction patterns from these experimental 3D structures. Different other automobile learning techniques, deep neural networks concur the promise of learning from millions of data without reaching a performance plateau speedily, which is computationally tractable by harvesting hardware accelerators (such every bit GPUs, TPUs) and parallel file system technologies. Wang et al5. have trained 3D deep convolutional networks (CNNs) on 3D grids representing poly peptide-poly peptide interfaces to evaluate the quality of docking models (DOVE). Gaiza et al6. have recently practical Geodesic CNNs to extract poly peptide interaction fingerprints past applying 2d ConvNets on spread-out poly peptide surface patches (MaSIF). Graph Neural Networks (GNNs)8, representing poly peptide interfaces equally graphs, have also been applied to predict poly peptide interfacesvii. Finally, rotation-equivariant neural networks have recently been used by Eisman et al. on point-based representation of the protein atomic construction to allocate PPIsnine. One outstanding illustration of the potential of deep neural networks in structural biological science is the recent quantum in single-chain poly peptide structure predictions by AlphaFold210,11,12 in the latest CASP14 (Critical Cess of protein Structure Prediction round xiv). Predicting the 3D structure of protein complexes remains however an open claiming: in CASP14 no single assembly was correctly predicted unless a known template was available. This calls for open-source frameworks that tin exist easily modified and extended by the community for data mining protein complexes and can expedite knowledge discovery on related scientific questions.
Data mining 3D poly peptide complexes presents several unique challenges. First, poly peptide interfaces are governed past physico-chemical rules. Different types of poly peptide complexes (e.grand., enzyme-substrate, antibody-antigen) may have different dominant interaction signatures. For case, some complexes may exist driven by hydrophobicity, and others by electrostatic forces. Second, protein interactions tin can be characterized at different levels: Atom-atom level, remainder-residue level, and secondary construction level. Third, protein interfaces are highly various in terms of shapes, sizes, and surface curvatures. Finally, efficient processing and featurization of a large number of atomic coordinates files of proteins is daunting in terms of computational cost and file storage requirements. There is therefore an emerging need for generic and extensible deep learning frameworks that scientists can easily re-use for their item problems, while removing tedious phases of information preprocessing. Such generic frameworks take already been developed in diverse scientific fields ranging from computational chemical science (DeepChem13) to condensed matter physics (NetKetxiv) and have significantly contributed to the rapid adoption of auto learning techniques in these fields. They take stimulated collaborative efforts, generated new insights, and are continuously improved and maintained past their respective user communities.
Here we innovate DeepRank, a generic deep learning platform for data mining protein-protein interfaces (PPIs) based on 3D CNNs. DeepRank maps diminutive and residue-level features calculated from 3D atomic coordinates of biomolecular complexes in Protein Data Bankfifteen (PDB, www.wwpdb.org) format onto 3D grids. DeepRank applies 3D CNN on these grids to learn problem-specific interaction patterns for user-defined tasks. The compages of DeepRank is highly modularized and optimized for high computational efficiency on very large datasets upwardly to millions of PDB files. It allows users to define their own 3D CNN models, features, target values (e.grand., class labels), and data augmentation strategy. The platform can be used both for classification, e.g., predicting an input PPI equally biological or a crystal artifact, and regression, due east.g., predicting binding affinities.
In the following, we first draw the structure of our DeepRank framework. To demonstrate its applicability and potential for structural biology, we employ it to 2 unlike inquiry challenges. We first nowadays the operation of DeepRank for the nomenclature of biological vs. crystallographic PPIs. With an accurateness of 86%, DeepRank outperforms country-of-the-art methods, such as PRODIGY-crystal16,17 and PISA18, which respectively attain an accuracy of 74 and 79%. We then present the performance of DeepRank for the scoring of models of protein-poly peptide complexes generated by computational docking. We show hither that DeepRank is competitive and sometimes outperforms 3 state-of-the-art scoring functions: HADDOCKxix, iScorexx,21, and DOVE5.
Results
Description of DeepRank
DeepRank is built every bit a Python 3 package that allows end-to-end training on datasets of 3D protein-protein complexes. The overall architecture of the package can be constitute in Supplementary Annotation 1 together with details regarding its implementation. The framework consists of 2 chief parts, one focusing on data pre-processing and featurization and the other on the preparation, evaluation, and testing of the neural network. The featurization exploits MPI parallelization together with GPU offloading to ensure efficient computation over very big data sets.
Data pre-processing and featurization:
- 1.
Feature calculations. Starting from the PDB files that describe the 3D structures of protein-protein complexes, DeepRank leverages pdb2sql22, our fast and versatile PDB file parser using Structured Query Language (SQL), to identify interface residues betwixt the 2 bondage. Interface residues are by default defined equally those with any atoms within a 5.5 Å distance cutoff (configurable) from any atom of another chain (Fig. 1A). The diminutive and residuum-based features presented in Table i are by default calculated, just users can easily define new features and include them in their feature calculation workflow.
Fig. 1: The DeepRank framework. 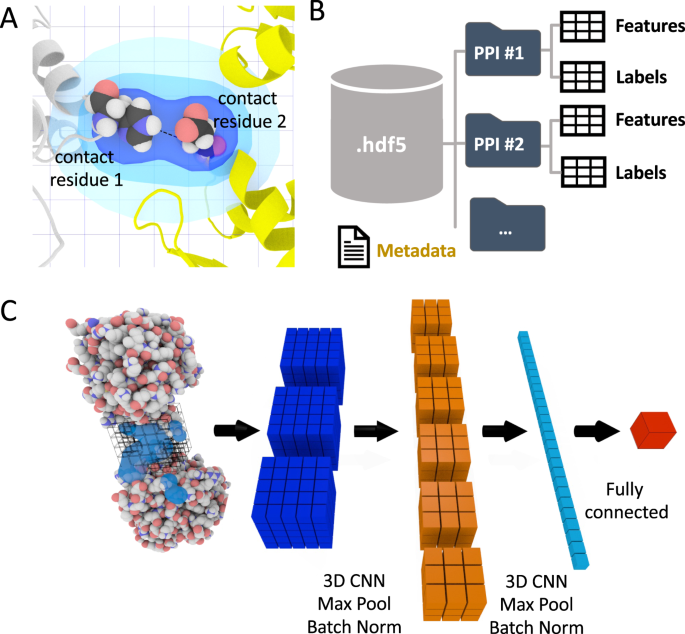
A The interface definition used by DeepRank. A residue is considered an interface residue if it is within a distance cutoff (v.5 Å by default, adjustable) of any atom on the other chain. The properties of interface residues or their atoms are used as features mapped on a 3D grid centered onto the interface. B Efficient storage of protein coordinates, features, and labels in HDF5 files. Given PDB files of protein-poly peptide complexes, DeepRank determines interface residues, calculates features, and maps the features onto 3D grids, storing these information, along with necessary metadata into HDF5 files. These files are the input for the subsequent CNN (Convolutional Network). This HDF5 format profoundly facilitates and speeds upward the retrieval of specific information. C Illustration of the training procedure. Users may select a subset of features and/or a subset of PPIs (protein-protein interfaces) contained in the HDF5 file and use them as input for a 3D CNN. The example network consists of several layers that mix convolution, max pooling, batch norm operations too every bit fully continued layers. The output of the network is the prediction of user-defined targets. Both nomenclature and regression are supported.
Table 1 Interface features predefined in DeepRank. - ii.
3D grid feature mapping. DeepRank maps the atomic and residue features of the interface of a complex onto a 3D grid using a Gaussian mapping (see Methods). The grid size and resolution can exist adjusted by users to suit their needs. Fig. 1A illustrates the mapping process for a residue-based feature (see the Methods section for explanations). Cheers to this gaussian mapping, each feature has a non-local upshot on the 3D characteristic filigree, contributing to a multitude of grid points. This feature mapping of the PPIs results in a 3D image where each grid point contains multiple channel values corresponding to different properties of the interface. Several data augmentation and PPIs structure alignment strategies are available to enrich the dataset.
- three.
Flexible target value definitions and calculations. Users may easily define problem-specific target values for their protein structures. For the scenario of computational docking, standard metrics to evaluate the quality of a docking model, i.east., by comparison to a reference construction, as used in the CAPRI (Critical Cess of PRedicted Interactions)23, are integrated into DeepRank. These include ligand RMSD (Root Hateful Foursquare Deviation)24, interface RMSD (iRMSD)24, FNAT (Fraction of Native Contacts)24, CAPRI quality labels24, and DockQ score25. DeepRank leverages pdb2sql22 to perform these calculations efficiently.
- 4.
Efficient data storage in HDF5 format. Dealing with sometimes tens of millions of small-scale-size PDB files with rich feature representations presents a claiming both for the file arrangement and for efficient preparation of deep neural networks. DeepRank stores the characteristic grids in HDF5 format, which is especially suited for storing and streaming very large and heterogeneous datasets. The general structure of the HDF5 generated past DeepRank is represented in Fig. 1B.
To train the neural network, DeepRank relies on the popular deep learning framework PyTorch26. The full general network architecture used in this piece of work is illustrated in Fig. 1C. Starting from the HDF5 files, users tin can easily select which features and target value to use during training and which PPIs to include in the training, validation, and test sets. It is as well possible to filter the PPIs based on their target values, for example by merely using docking models with an iRMSD values to a higher place or below a certain threshold, thus discarding unrealistic data points. The input data are fed into a series of 3D convolutional layers, max pool layers, and batch normalization layers, ordinarily followed by fully connected layers. The exact architecture of the network as well every bit all other hyper parameters tin exist easily modified past users to tune the grooming for their particular applications (run across Supplementary Notes ane and 4). The result of the grooming is stored in a dedicated HDF5 file for subsequent analysis.
Application 1: Detecting crystal artifacts
X-Ray crystallography is one of the nearly of import experimental approaches to determine 3D structures of protein complexes (information technology accounts for >80% of all deposited PDB entries). This experimental technique kickoff requires the proteins to be crystallized and then exposed to X-rays to obtain their structures. When information technology comes to structures of complexes, the resulting crystals oft incorporate multiple interfaces, some of which are biologically relevant and some are mere artifacts of the crystallization process, the so-chosen "crystal interfaces" (Fig. 2A, B). Distinguishing crystal interfaces from biological ones, when no additional information is available, is still challenging. Several computational approaches have been proposed to distinguish such interfaces, among which PISAxviii and PRODIGY-crystalsixteen,17 testify the highest prediction performances. PISA is based on six physicochemical properties: Costless energy of formation, solvation energy gain, interface area, hydrogen bonds, salt-span across the interface, and hydrophobic specificity. PRODIGY-crystal is a random woods classifier based on structural backdrop of interfacial residues and their contacts16.
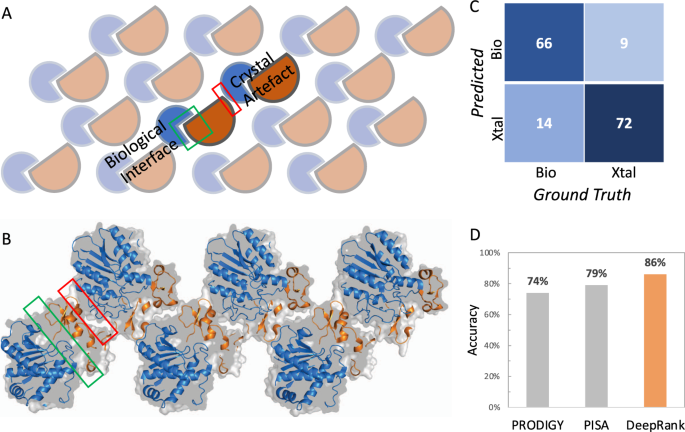
A, B. Illustration of the two types of interfaces, i.due east., biological and crystal interfaces, found in a crystal. Protein molecules are orderly arranged in repetitive crystal units. Crystallographic interfaces tin can originate from the seeming interaction from the two neighboring crystal units, which may or may not represent biological interactions. The crystal structure shown in B corresponds to PDB entry 1ZLH. C Confusion matrix of DeepRank on the DC test dataset. D Accurateness of DeepRank, PISA, and PRODIGY-crystal on the DC dataset.
We applied DeepRank to the problem of classifying biological vs. crystal interfaces. Nosotros trained and validated the 3D CNN specified in Supplementary Notation three on the MANY dataset27, which consists of 2828 biological interfaces and 2911 crystal ones, only using Position Specific Scoring Matrix (PSSM) features. Each structure was first augmented by random rotation (thirty times) earlier training. Early stopping on the validation loss was used to determine the optimal model (see Supplementary Fig. 3). The trained network was tested on the DC dataset28, containing lxxx biological and 81 crystal interfaces. On this examination set up, the trained network correctly classified 66 out of 80 biological interfaces and 72 out of 81 crystal interfaces (Fig. 2C). DeepRank thus accomplished an accuracy of 86%, outperforming PRODIGY-crystal and PISA, which reported 74 and 79%, respectively16 (Fig. 2D). While 89 test cases present at least one homolog in the MANY dataset, removing these cases from the testing dataset even so leads to satisfying performance with an accuracy of 82%. (Supplementary Table 1).
Application two: Ranking docking models
Computational docking is a valuable tool for generating possible 3D models of poly peptide complexes and provides a complementary alternative to experimental structure determination. Given the 3D structures of individual proteins, docking aims at modeling their interaction mode past generating typically tens of thousands of candidate conformations (models). Those models are ranked using a scoring role to select the correct (almost-native) ones (Fig. 3A). Although much attempt is dedicated to amend the scoring23,29,thirty,31, reliably distinguishing a native-similar model from the vast number of incorrectly docked models (wrong models) remains a major challenge in docking.
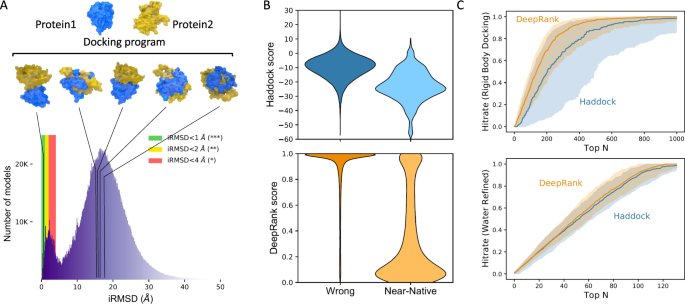
A Acme: Using a docking software (east.g., HADDOCK36) a large number of docking poses between 2 proteins (here PDB ID: 1AK4) are generated. Lesser: Distribution of iRMSD (interface root hateful squared divergence) values obtained for the docking criterion 532 using HADDOCK in 5 docking scenarios (meet Methods). Well-nigh conformations (93%) are incorrect with iRMSD values larger than 4 Å and merely less than 1% of the conformations have iRMSD values lower than i Å. B Distribution of the DeepRank and HADDOCK scores for the wrong models and near-native models from the rigid body docking phase. The lower the score the higher likelihood a model is predicted to be a almost-native model. C Performance of DeepRank on the BM5 set compared to HADDOCK's scoring role. This data represents the predictions of both methods on 140 distinct examination cases considered during the ten-fold cross-validation. Each individual test case contains about 3000 conformations of a single complex. The thick line marks the median of the Hit Rates (see Methods) calculated for individual exam cases and the shaded surface area marks the 25–75% quantile interval. The data is shown up to the top chiliad (encounter Supplementary Fig. half-dozen for the full-range plot). Top: Rigid-trunk docking models merely; Bottom: Water refined models simply. HADDOCK uses different scoring functions for models generated in dissimilar stages: rigid-body, flexible-docking, and h2o-refinement stages (encounter Methods).
Nosotros used HADDOCK19 to generate a fix of docking models of various qualities for the docking criterion v5 (BM5) ready32, including both rigid-body docking, flexible docking, and final refined docking models. In this work, we focused on 142 dimers for which near-native models were available in the generated data sets, excluding all antibody-antigen complexes.
We trained the 3D CNN (architecture specified in Supplementary Note three) using all available atomic and residue-based features (Tabular array 1), mapped onto a filigree of 30 × xxx × 30 Åiii with 1 Å resolution. The network was trained on over 300,000 labeled docking conformations to classify models as about-native or wrong. The DeepRank score, i.e., the predicted likelihood of a model to be a wrong model, was then used to rank the models for each example. To ensure objective evaluations, we conducted x-fold cross-validation at the level of complexes, i.e., all models for a given complex are used exclusively in either training, validation, or test sets in each fold.
DeepRank performs well on HADDOCK models generated in the rigid-body docking stage. The DeepRank scores are well separated between near-native and incorrect models (Fig. 3B), while the HADDOCK scores present a significant overlap between those two classes. In terms of ranking functioning, DeepRank outperforms HADDOCK on rigid-trunk docking models by selecting more near-native models among Peak N (Fig. 3C top). The narrow spread of the values obtained with DeepRank, illustrated by the 25–75% quantile interval, indicates that DeepRank is rather consistent in its ranking of different cases, while HADDOCK presents poor operation for some cases. This divergence might be explained by the fact that DeepRank is less sensitive to the detailed energetics of the interface than the HADDOCK score.
The differences between DeepRank and HADDOCK are less pronounced for water-refined docking models (Fig. 3C bottom). However, note that HADDOCK requires using different scoring functions for models generated in rigid-trunk, flexible-docking, and water-refinement stage while DeepRank employ the aforementioned scoring function for all stages (see Methods and Supplementary Fig. six). An assay of Success Rate of DeepRank and HADDOCK at the unlike stages (Supplementary Fig. 7) confirms the expert operation of DeepRank that slightly outperforms HADDOCK for each model type. This confirms again the robustness of the DeepRank score, since it provides a unmarried score that performs well beyond differently refined models.
To further examination the operation of DeepRank we have trained a terminal 3D CNN model using the docking conformations of all the 142 BM5 dimer complexes and practical it to 13 cases from the CAPRI score fix33. We compared DeepRank with three leading scoring functions, the HADDOCKnineteen scoring function that uses an energy-based arroyo; the recently adult iScorexx,21 a graph-kernel based scoring function; and Pigeon5 a contempo deep-learning method besides based on 3D CNNs. DeepRank is competitive with these scoring functions, fifty-fifty outperforming them on some cases (Supplementary Fig. eight and Supplementary Table two). Our results also suggest the ability of DeepRank to correctly identify favorable interactions that are ignored by the other methods, which might betoken a possible complementarity of these approaches (Supplementary Figs. 9 and 10).
Discussion
We have presented here our DeepRank framework, demonstrating its use and operation on 2 structural biological science challenges. Its main advantages are as follows:
- 1.
From a user's perspective, the platform provides a user-friendly interface. Information technology implements many options that tin exist easily tuned. Information technology provides flexibility through the featurization and the design of the neural network architecture (see code snippets in Supplementary Note iv). This makes it directly applicative for a range of problems that use protein-poly peptide interfaces every bit input information.
- 2.
From a programmer's perspective, DeepRank is adult equally a software package following software development standards34 including version control, continuous integration, documentation, and easy addition of new features. This flexibility increases the maintainability and further development of DeepRank by the community, for example, to allow predicting mutation effects on unmarried protein structures.
- 3.
Computational efficiency: in all stages, DeepRank has been developed to make it possible to use millions of PDB files to train models, and examination their performance.
- iv.
Finally, the performances competing and outperforming the country-of-the-art on two different research bug demonstrate the versatility of DeepRank in general structural biological science.
When applied to the classification of biological versus crystallographic interfaces (awarding one), the trained network (provided in Information Availability) shows satisfying performance leading to a improve classification than competing methods, PRODIGY-crystal and PISA. This comeback is due to the use of evolution information through the PSSM and from the apply of deep neural network that are capable of learning the subtle differences between the interaction patterns of the 2 types of interfaces.
When because the scoring problem (awarding 2), DeepRank performs specially well on scoring rigid-body HADDOCK models, outperforming the HADDOCK's rigid-body-specific Score (Fig. 3C). Since rigid-body docking is the showtime modeling stage of HADDOCK, incorporating DeepRank scoring in the HADDOCK docking workflow is expected to amend the modeling success rate past passing more high-quality models to the later flexible refinement stages. This event also indicates that our trained network (provided in Data Availability) could exist generally applicative to models from a variety of rigid-body docking software. DeepRank is robust on different type of models (rigid-trunk, flexible-refined, water-refined) (Fig. 3C) and not sensitive to clashes at the interface. This broad applicability range is of import in experiments similar the community-broad CAPRI scoring experiment where a mixture of highly refined and rigid-body models (that often nowadays unphysical diminutive arrangements, or clashes) take to be scored23. While DeepRank does not significantly outperform HADDOCK iScore nor DOVE on selected cases from previous rounds of CAPRI, it does identify different conformations as being near-native (Supplementary Fig. 9), indicating a complementarity in scoring. The comparison of the dissimilar methods clearly illustrates the difficulty in obtaining a model that performs consistently across the diversity of PPIs and calls for more research to engineer meliorate featurization, datasets, and scoring functions.
Currently DeepRank converts irregular-shaped PPIs into structured information (i.e., 3D grids) and so that 3D CNN tin be applied. These structured 3D grids could besides be used with equivariant neural networks35 that naturally incorporate translation- and rotation-invariance and hence avoids the data augmentation that is sometimes needed when using 3D CNN. The utilize of non-structured geometric information such as graphsvii, surfaceshalf-dozen, or betoken clouds as input, offer additional opportunities for the futurity development of DeepRank. For example, MaSIF6 exploits geodesic CNN to extract protein interaction fingerprint and therefore just requires information augmentation in 2nd instead of 3D. Yet, the information preprocessing required by MaSIF to determine protein surface patches, calculate polar coordinates and map the features, is nigh 48 times more computationally demanding and 7 times more retention demanding than calculating all the 3D grids required by DeepRank (see Supplementary Table 3). This hinders the applicability of MaSIF to large-calibration analyses on millions of protein models obtained for example in computational docking or large-scale modeling of mutations. Even so, because the potential of geometric learning with respect to rotation-invariance, it would exist useful to extend DeepRank with geometric deep learning techniques to more efficiently stand for PPIs with highly irregular shapes. Some other enhancement would exist to extend the framework to handle complexes containing more than than two bondage to broaden its application scope.
In summary, we accept described an open-source, generic, and extensible deep learning framework for data mining very large datasets of protein-protein interfaces. We demonstrated the effectiveness and readiness of applying DeepRank on two dissimilar challenges in structural biology. We expect DeepRank to speed-up scientific inquiry related to poly peptide interfaces by facilitating the boring steps of data preprocessing and reducing daunting computational costs that may be associated with large-calibration information analysis. Its modularized and extendable framework bears slap-up potential for stimulating collaborative developments by the computational structural biology community on other protein structure-related topics and will contribute to the adoption and evolution of deep learning techniques in structural biology research.
Methods
Gaussian mapping of the atomic and residue features
The diminutive and residual features are mapped on a 3D grid using Gaussian functions. The contribution w k of cantlet k (or residue) to a given grid point follows a hence a Gaussian distance dependence:
$${w}_{k}(r)={five}_{yard}{exp} (||r-{r}_{k}|{|}^{2}/ii{\sigma }^{2})$$
(1)
where v k is the value of the feature, r k the (x,y,z) coordinate of cantlet m, and r the position of the filigree point. For atomic features the standard deviation σ is taken equally the van der Waals radius of the corresponding atom. In instance of residue-based features, r grand is taken as the position of the blastoff-carbon atom of the residue and σ as the van der Waals radius of that cantlet.
For pairwise features, such as interaction energy terms, the value of v k is defined as the sum of the all the interaction terms between atom (residuum) k and its contact atoms(residues): \({5}_{k}={\sum }_{l}{v}_{kl}\) where five kl is the interaction term between atoms(residues) k and l.
Application 1: Detecting crystal artifacts
Information Sets: The original MANY dataset27 consists of 2831 biological interfaces and 2912 crystal ones. Entries 2uuy_1, 2p06_1 and 3dt5_1 were excluded due to all zero values of PSSM information content (IC) and 1gc2_5 was excluded due to failed residuum density calculation. We thus used 2828 biological interfaces and 2911 crystal ones, which was used to create the training (80%) and validation (20%) sets while maintaining the balance between positive and negative data. The training set up was then further augmented past randomly rotating each complex 30 times. The DC dataset28 was used every bit a test ready. It contains fourscore biological and 81 crystal interfaces, excluding 3jrz_1 due to failed residue density adding. All entries of both the MANY and the DC dataset accept been previously refined16 using the refinement protocol of our HADDOCK web server36.
Grooming and evaluations: The architecture of the CNN used in our experiments is shown in Supplementary Fig. 2. Nosotros used the cantankerous-entropy and log softmax as loss and scoring functions, respectively, and stochastic slope descent equally optimizer. The learning charge per unit was set to 0.0005, momentum to 0.nine, weight decay to 0.001, and batch size to 8. Equally features we used only the PSSM every bit evolutionary information, which has been demonstrated to be useful for charactering poly peptide interfaces4. Nosotros used 10 × x × 10 Å3 grids with a resolution of 3 Å. This low-resolution grid is sufficient to capture the variation of the PSSM which are residue-based features and thus less finely divers than diminutive-based features.
Application 2: Ranking docking models
Data sets: The dataset for scoring was generated using HADDOCK, an integrative modeling softwarenineteen. Docking in HADDOCK follows three stages: (1) it0: rigid-body docking, (ii) it1: semi-flexible refinement by simulated annealing in torsion angle infinite, and (3) itw: final refinement past curt molecular dynamics in explicit solvent (default: water). Nosotros used HADDOCK37 to systematically generate a ready of docking models for the docking criterion v5 set32, BM5. In club to generate a suitable amount of most-native models both guided and ab-initio dockings were performed following 5 different scenarios: (1) Refinement of the bound complexes (50/50/l models for it0/it1/water stages, referred to as "refb"), (ii) guided docking using true interface divers at 3.nine Å cutoff every bit cryptic interaction restraints (1000/400/400 models for it0/it1/water, "ti"), (3) guided docking using true interface defined at 5.0 Å cutoff (1000/400/400 models for it0/it1/h2o, "ti5"), (4) ab-initio docking with center of mass restraints (10,000/400/400 models for it0/it1/water, "cm"), and (v) ab-initio docking with random surface patch restraints (ten,000/400/400 models for it0/it1/water, "ranair"). Scenarios ii–5 are unbound-unbound docking, providing existent-life challenges (i.eastward., conformational changes upon bounden) to DeepRank. BM5 consists of 232 non-redundant cases (the non-redundancy was here evaluated at the SCOP family level32). In total, nosotros generated ~5.84 million HADDOCK models (25,300 models per case) corresponding to 11 TB of data (3D coordinates plus characteristic grids) in HDF5 format. In this study, we focused on 142 dimers, i.e., cases with ii bondage (1IRA was excluded due to the lack of PSSM for the short concatenation A).
As the models were not clustered, a big degree of redundancy exists in the dataset, with very similar conformations being represented multiple times. After experimentation, nosotros observed that because only a subset of about 420 M models (~3000 models per complex) was sufficient to accurately represents the available information (see Supplementary Fig. iv for details). Most 30 K models were near-native models among these 420 K models. Each docking model was randomly rotated to limit the sensitivity of our results to a particular orientation of the PPIs.
Network architectures and weight optimizations: The compages of the CNN in our experiment is shown in Supplementary Fig. v. We used the cross-entropy loss function over the raw scoring output. Nosotros used the Adam optimizer with a learning rate of 0.001. The batch size during training was 100. We used the full set up of 36 physico-chemic features (channels) that are predefined in DeepRank (Tabular array i). We used a grid size was 30 x xxx x 30 Å3 with a resolution of 1 Å to accurately capture the variation of diminutive-based features and to adequately resolve the average cached surface surface area per monomer (BSA) of a dimer PPI in the BM5 set, that is virtually 909 Å2. We evaluate the BSA as:
$${{{{{\rm{BSA}}}}}}=\frac{one}{2}({{{{{{\rm{ASA}}}}}}}_{{{{{{\rm{chainA}}}}}}}+{{{{{{\rm{ASA}}}}}}}_{{{{{{\rm{chainB}}}}}}}-{{{{{{\rm{ASA}}}}}}}_{{{{{{\rm{complex}}}}}}})$$
(two)
where ASA stands for the accessible surface area.
Considering our dataset is highly imbalanced (only around 7.one% of the models are positives, i.due east., with iRMSD 4 Å), for training the network nosotros assigned class weights to the loss office, proportional to the form sizes, that is, the class weights are 0.071 and 0.929 for the negatives and positives, respectively so that the loss role penalizes equally errors made on the positive and on the negative dataset.
Training, validation, and testing: We performed a 10-fold cross-validation on the BM5 set. In every fold, out of 142 complexes, we used models from 114 complexes for preparation, 14 complexes for validation, and xiv complexes were reserved for testing. In this way, in total 140 out of the 142 complexes were used in the test sets (2 complexes BAAD and 3F1P are not used in the testing to proceed the testing set having an equal number of complexes for each fold). The grooming, validation, and testing sets are disjunctive at complex-level so that complexes in the testing ready are never seen by the neural networks during its training and validation. In the stop, we trained 1 network for each fold (thus in full 10 trained networks for ten-CV), and the set-aside independent examination set was evaluated on this network.
The network is trained to perform positive/negative classification of models, outputting two scores equally predicted probability of an input model being virtually-native or wrong. We and so ranked the docking models based on their predicted scores of being wrong models to be consequent with widely-used energy-based scoring functions: i.e., a low prediction score indicates that the function predicts this model is of high quality. For the final analysis (Supplementary Fig. 7), we merged prediction results from 10 folds in our performance reports on the BM5 gear up.
To evaluate DeepRank on the CAPRI score prepare, we take start trained one network on all the 449,158 docking models from 142 BM5 complexes. This trained neural network model was then used on the CAPRI score fix33.
HADDOCK scoring functions: HADDOCK uses three different scoring functions for models generated at different docking stages. We neglected the AIR restraints to facilitate the comparing between the different model qualities. We used the following functions:
$${{{{{{\rm{HADDOCK}}}}}}}_{{{{{{\rm{information technology}}}}}}0}=0.01{{{{{{\rm{E}}}}}}}_{{{{{{\rm{vdw}}}}}}}+1.0{{{{{{\rm{E}}}}}}}_{{{{{{\rm{elec}}}}}}}+1.0{{{{{{\rm{East}}}}}}}_{{{{{{\rm{desol}}}}}}}-0.01{{{{{\rm{BSA}}}}}}$$
(3)
$${{{{{{\rm{HADDOCK}}}}}}}_{{{{{{\rm{it}}}}}}ane}=1.0{{{{{{\rm{E}}}}}}}_{{{{{{\rm{vdw}}}}}}}+1.0{{{{{{\rm{Due east}}}}}}}_{{{{{{\rm{elec}}}}}}}+1.0{{{{{{\rm{E}}}}}}}_{{{{{{\rm{desol}}}}}}}-0.01{{{{{\rm{BSA}}}}}}$$
(4)
$${{{{{{\rm{HADDOCK}}}}}}}_{{{{{{\rm{itw}}}}}}}=1.0{{{{{{\rm{E}}}}}}}_{{{{{{\rm{vdw}}}}}}}+0.ii{{{{{{\rm{E}}}}}}}_{{{{{{\rm{elec}}}}}}}+1.0{{{{{{\rm{E}}}}}}}_{{{{{{\rm{desol}}}}}}}$$
(5)
where HADDOCKit0, HADDOCKit1, and HADDOCKitw are the scoring functions used respectively for rigid-body docking, semi-flexible docking and water-refined models. Evdw is the van der Waals intermolecular free energy, Eelec the electrostatic intermolecular energy, Due eastdesol the desolvation energy and BSA the buried surface surface area.
Evaluation metrics: Hit Rate and Success Charge per unit are used to evaluate the performance of scoring functions. The Hit Rate is defined as the pct of hits (models with iRMSD ≤4 Å) in the top ranked models for a specific complex:
$${{{{{\rm{Hit}}}}}}\,{{{{{\rm{Rate}}}}}}(M)=\frac{{due north}_{{{{{{\rm{hits}}}}}}}(K)}{M}$$
(6)
where due north hits(1000) is the number of hits (i.e., almost-native models) among height models and G the full number of near-native models for this case. The Hit Rate was calculated for each individual case in our exam set and statistics across the different cases (median, 1st and 3rd quartile) were calculated. As the total number of models varies between cases, these statistical values were only evaluated from K = 1 to K =Northward min where N min is the smallest number of models that all cases have.
The Success Rate shown in Supplementary Fig. vii is defined as the per centum of complexes for which at least one near-native model is found in the tiptop N selected models. Information technology is therefore divers as:
$${{{{{\rm{Success}}}}}}\,{{{{{\rm{Rate}}}}}}=\frac{{north}_{{{{{{\rm{successful}}}}}}\_{{{{{\rm{cases}}}}}}}(K)}{N}$$
(7)
where northward successful_cases(K) is the number of cases with at least one near-native model amongst top models, and North is the full number of cases.
Reporting summary
Farther data on enquiry blueprint is available in the Nature Enquiry Reporting Summary linked to this article.
Data availability
The PDB files and PSSM files for the two experiments along with the lawmaking for training and post analysis have been deposited in SBGrid (information.sbgrid.org, https://doi.org/ten.15785/SBGRID/843).
Code availability
The DeepRank software has been released to the Python Bundle Alphabetize at https://pypi.org/projection/deeprank/ (https://doi.org/10.5281/zenodo.3735042)38. Its source code and documentation are freely bachelor at https://github.com/DeepRank/deeprank and https://deeprank.readthedocs.io, respectively. The PSSMs used in this paper were calculated using our PSSMgen packet: https://github.com/DeepRank/PSSMGen (https://doi.org/x.5281/zenodo.4509544)39.
References
-
Gane, P. J. & Dean, P. Yard. Recent advances in structure-based rational drug design. Curr. Opin. Struct. Biol. x, 401–404 (2000).
-
Sadelain, M., Brentjens, R. & Rivière, I. The bones principles of chimeric antigen receptor design. Cancer Discov. iii, 388–398 (2013).
-
Liu, S. et al. Nonnatural protein–protein interaction-pair design past key residues grafting. Proc. Natl. Acad. Sci. 104, 5330–5335 (2007).
-
Xue, L. C., Dobbs, D., Bonvin, A. One thousand. J. J. & Honavar, V. Computational prediction of poly peptide interfaces: a review of data driven methods. FEBS Lett 589, 3516–3526 (2015).
-
Wang, X., Terashi, Grand., Christoffer, C. W., Zhu, M. & Kihara, D. Protein docking model evaluation by 3D deep convolutional neural networks. Bioinformatics 36, 2113–2118 (2020).
-
Gainza, P. et al. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 17, 184–192 (2020).
-
Fout, A., Byrd, J., Shariat, B. & Ben-Hur, A. Poly peptide interface prediction using graph convolutional networks. in Proc. 31st International Conference on Neural Information Processing Systems 6533–6542 (Curran Associates Inc., 2017).
-
Z, W. et al. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Larn. Syst. https://doi.org/10.1109/tnnls.2020.2978386 (2020).
-
Eismann, S. et al. Hierarchical, rotation‐equivariant neural networks to select structural models of poly peptide complexes. Proteins Struct. Funct. Bioinforma 89, 493–501 (2021).
-
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
-
Jumper, J. et al. High Accurateness Poly peptide Structure Prediction Using Deep Learning. CASP 14 Abstr. Book (2020).
-
Jumper, J. et al. Highly authentic protein structure prediction with AlphaFold. Nature 1–xi https://doi.org/ten.1038/s41586-021-03819-ii (2021).
-
Ramsundar, B., Eastman, P., Walters, P. & Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More. (O'Reilly Media, 2019).
-
Carleo, G. et al. NetKet: a auto learning toolkit for many-body quantum systems. SoftwareX 10, 100311 (2019).
-
Berman, H. M. et al. The protein information bank. Nucleic Acids Res. 28, 235–242 (2000).
-
Jiménez-García, B., Elez, Thousand., Koukos, P. I., Bonvin, A. M. & Vangone, A. PRODIGY-crystal: a spider web-tool for classification of biological interfaces in protein complexes. Bioinformatics 35, 4821–4823 (2019).
-
Elez, K., Bonvin, A. M. J. J. & Vangone, A. Distinguishing crystallographic from biological interfaces in protein complexes: role of intermolecular contacts and energetics for classification. MC Bioinform. 19, 438 (2018).
-
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
-
Dominguez, C., Boelens, R. & Bonvin, A. Yard. J. J. HADDOCK: a protein−poly peptide docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
-
Geng, C. et al. iScore: a novel graph kernel-based function for scoring protein–protein docking models. Bioinformatics 36, 112–121 (2020).
-
Renaud, N. et al. iScore: an MPI supported software for ranking poly peptide–protein docking models based on a random walk graph kernel and support vector machines. SoftwareX 11, 100462 (2020).
-
Renaud, N. & Geng, C. The pdb2sql Python bundle: parsing, manipulation and analysis of PDB files using SQL queries. J. Open Source Softw. 5, 2077 (2020).
-
Lensink, M. F. & Wodak, S. J. Docking and scoring protein interactions: CAPRI 2009. Proteins Struct. Funct. Bioinforma 78, 3073–3084 (2010).
-
Méndez, R., Leplae, R., Maria, L. D. & Wodak, S. J. Assessment of blind predictions of protein–protein interactions: current status of docking methods. Proteins Struct. Funct. Bioinforma 52, 51–67 (2003).
-
Basu, S. & Wallner, B. DockQ: a quality measure for protein-protein docking models. PLOS I 11, e0161879 (2016).
-
Paszke, A. et al. PyTorch: An imperative style, high-functioning deep learning library. in Advances in Neural Information Processing Systems 32 (eds. Wallach, H. et al.) 8026–8037 (Curran Associates, Inc., 2019).
-
Baskaran, Grand., Duarte, J. M., Biyani, N., Bliven, S. & Capitani, G. A PDB-wide, development-based assessment of protein–poly peptide interfaces. BMC Struct. Biol. 14, 22 (2014).
-
Duarte, J. M., Srebniak, A., Schärer, M. A. & Capitani, Grand. Poly peptide interface classification by evolutionary analysis. BMC Bioinformatics 13, 334 (2012).
-
Gromiha, M. M., Yugandhar, Grand. & Jemimah, S. Poly peptide–protein interactions: scoring schemes and binding affinity. Curr. Opin. Struct. Biol. 44, 31–38 (2017).
-
Geng, C., Xue, L. C., Roel‐Touris, J. & Bonvin, A. 1000. J. J. Finding the ΔΔG spot: are predictors of binding affinity changes upon mutations in poly peptide–protein interactions ready for it? WIREs Comput. Mol. Sci. 9, e1410 (2019).
-
Zhang, Q. et al. Recent advances in protein-protein docking. Curr. Drug Targets 17, 1586–1594 https://www.eurekaselect.com/138443/article (2016).
-
Vreven, T. et al. Updates to the integrated protein–protein interaction benchmarks: docking benchmark version 5 and analogousness benchmark version 2. J. Mol. Biol. 427, 3031–3041 (2015).
-
Lensink, M. F. & Wodak, S. J. Score_set: a CAPRI benchmark for scoring protein complexes. Proteins Struct. Funct. Bioinforma 82, 3163–3169 (2014).
-
Drost, Niels et al. Netherlands eScience Center—Software Development Guide. (Zenodo, 2020). https://doi.org/x.5281/ZENODO.4020565.
-
Smidt, T. Euclidean symmetry and equivariance in automobile learning. Trends Chem 3, 82–85 (2021).
-
van Zundert, G. C. P. et al. The HADDOCK2.2 spider web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016).
-
de Vries, S. J., van Dijk, M. & Bonvin, A. Chiliad. J. J. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 (2010).
-
Renaud, North. et al. DeepRank/deeprank v0.1.0. (Zenodo, 2020). https://doi.org/ten.5281/ZENODO.3735042.
-
Renaud, Nicolas & Geng, Cunliang. PSSMGen. (Zenodo, 2021). https://doi.org/10.5281/ZENODO.4509544.
-
Ragoza, M., Hochuli, J., Idrobo, East., Sunseri, J. & Koes, D. R. Protein–ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017).
-
Jorgensen, W. L. & Tirado-Rives, J. The OPLS (optimized potentials for liquid simulations) potential functions for proteins, energy minimizations for crystals of circadian peptides and crambin. J. Am. Chem. Soc. 110, 1657–1666 (1988).
-
Chothia, C. & Janin, J. Principles of poly peptide–protein recognition. Nature 256, 705–708 (1975).
-
Altschul, S. F. et al. Gapped Smash and PSI-Smash: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Acknowledgements
The project is supported past the ASDI grant provided by Netherlands eScience Center (Grant number ASDI.2016.043), past SURF Open Lab "Car learning enhanced HPC applications" grant (AB/FA/10573), and by a "Calculating Fourth dimension on National Computer Facilities" grant (2018/ENW/00485366) from NWO (Netherlands Organisation for Scientific Inquiry). AMJJB acknowledges financial support from the European Union Horizon 2020 projects BioExcel (675728, 823830) and EOSC-hub (777536). This piece of work was carried out on the Dutch national eastward-infrastructure with the support of SURF Cooperative (Grant ID: 2018/ENW/00485366). LX and DM acknowledge financial support by the Hypatia Fellowship from Radboudumc (Rv819.52706). Nosotros give thanks Dr. Valeriu Codreanu, Dr. Caspar van Leeuwen, and Dr. Damian Podareanu from SURFsara for providing HPC back up for efficient information processing using Cartesius, the Dutch national supercomputer.
Author data
Affiliations
Contributions
Fifty.Ten. and A.Grand.J.J.B. designed and supervised the project. L.10. contributed to the development and evaluations of DeepRank. A.Thou.J.J.B. generated the HADDOCK docking models. C.K., L.Ten., S.One thousand. and North.R. designed and adult the software. C.G. and F.A. performed the experiments for Awarding 1 (classification of biological interfaces vs. crystal ones). Southward.Thousand. performed the experiments for Awarding two (ranking docking models) and contributed to experiments in Application 1. D.M. contributed to writing the documentation and evaluations of DeepRank. MR contributed to the development of the software and the evaluation of the computational efficiency of DeepRank and MaSIF. L.R. organizes the project and contributed to the discussions and development of DeepRank. All authors contributed to the manuscript writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Nature Communications thanks Ilya Vakser and the other, anonymous, reviewer(south) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This commodity is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, accommodation, distribution and reproduction in any medium or format, as long equally you give appropriate credit to the original author(due south) and the source, provide a link to the Creative Commons license, and indicate if changes were fabricated. The images or other third party material in this article are included in the article's Artistic Commons license, unless indicated otherwise in a credit line to the material. If fabric is not included in the article's Creative Commons license and your intended utilize is not permitted past statutory regulation or exceeds the permitted use, y'all will demand to obtain permission straight from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
Nearly this article
Cite this article
Renaud, N., Geng, C., Georgievska, S. et al. DeepRank: a deep learning framework for information mining 3D protein-protein interfaces. Nat Commun 12, 7068 (2021). https://doi.org/10.1038/s41467-021-27396-0
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/s41467-021-27396-0
Comments
By submitting a comment yous concord to abide past our Terms and Customs Guidelines. If you detect something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Source: https://www.nature.com/articles/s41467-021-27396-0
0 Response to "Short Notes on the State of Art and Challenges on Data Mining"
Post a Comment